Circulating tumour DNA
Analyses of circulating tumour DNA (ctDNA) will improve response monitoring as it interrogates all tumours and can potentially measure tumour burden much more precisely than other modalities such as traditional imaging approaches. Changes in very small tumours may be invisible or undiscernible by imaging and traditional imaging approaches for immunotherapy have the disadvantage that immune and T cell activation can lead to unusual patterns of response on imaging, with the advent of so-called pseudo-progression, which refers to an increase of tumour burden and/or the appearance of new lesions due to infiltration by activated T cells before the disease responds to therapy (Dromain et al., 2020).
SAGAsafe® (formerly known as IBSAFE) will use digital PCR (dPCR) to detect and quantify point mutations with unique ultra-sensitivity to 0.001% MAF (Mutant Allele frequency). This is approximately 100-fold improved compared to other methods. The dPCR technique will allow for a fast turn-around with a possible result within hours if done in a local lab, or within a few days if sent to a remote lab. Compared to Next Generation Sequencing (NGS) it is also significantly less costly.
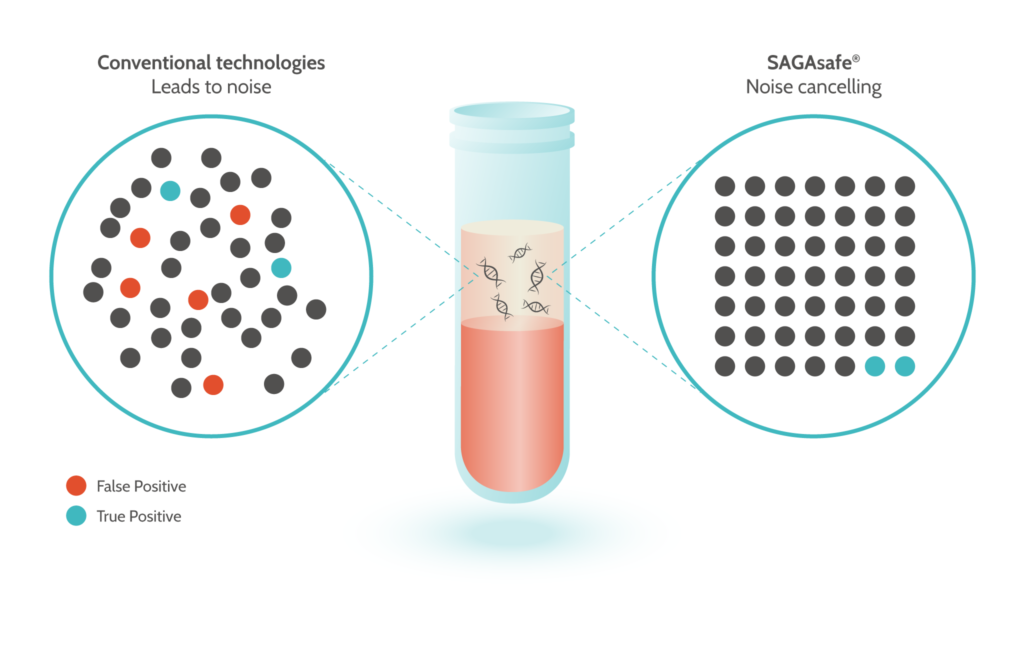
When monitoring cancer patients during our study, the technology will provide crucial quantitative information, potentially leading to breakthrough insights. The methods work equally well with liquid biopsies such as blood samples as well as on tissue biopsies.
Samples will be analysed before treatment (baseline) as well as during and after treatment. The amount of ctDNA to normal circulating DNA in a cancer patient varies considerably from near 100% to a fraction that is exceedingly low, e.g. from 0.1% down to 0.001% mutant allele fraction (MAF). Hence, to reliably analyse ctDNA, the ideal method of analysis would be able to detect an MAF as low as 0.001%. Among competing genomic methods, SAGAsafe® has an MAF limit of detection down to 0.001%. With this limit of detection, SAGAsafe® is able to detect true positives and represents a new state of the art.
There is growing evidence that circulating HPV DNA can be used as a marker for early treatment success. Recently, Rutkowski et al. (2020) showed that the post-treatment assessment of ctHPV16 DNA in patients with HPV-related oropharyngeal cancer may be used as a complementary biomarker to conventional imaging-based examinations for early identification of treatment failure.
